- Undergraduate
Bachelor's Degrees
Bachelor of ArtsBachelor of EngineeringDual-Degree ProgramUndergraduate AdmissionsUndergraduate Experience
- Graduate
Graduate Experience
- Research
- Entrepreneurship
- Community
- About
-
Search

Daniel Olson
Associate Professor of Engineering
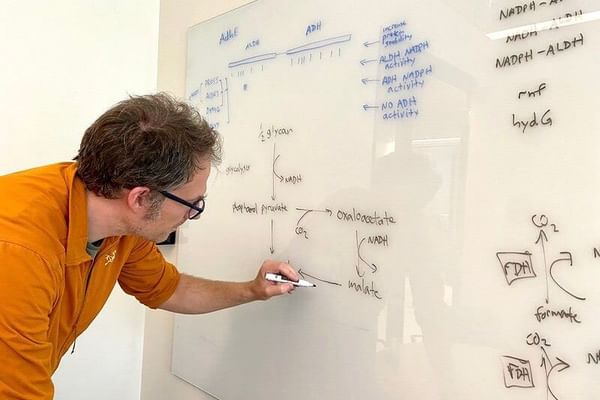
Professor Olson analyzes central metabolic pathways from Clostridium thermocellum. (Photo by Catha Mayor)
Research Interests
Metabolic engineering; cellulosic biofuel; metabolic flux analysis; genome resequencing analysis
Education
- BA, Physics, Dartmouth 2004
- BE, Engineering Sciences, Dartmouth 2006
- PhD, Engineering Sciences, Dartmouth 2011
Professional Activities
- Metabolic Engineering Task Leader, DOE Center for Bioenergy Innovation
- Biotechnology Focus Area Leader, Advanced Second Generation Biofuel (A2G) Laboratory, University of Campinas, Brazil
- Metabolic Engineering Group Leader, Lynd Lab
Research Projects
-
Genetic tools for anaerobic thermophilic bacteria
Genetic tools for anaerobic thermophilic bacteria
In large-scale industrial fermentations, it can be expensive to add oxygen, and to cool fermenters to mesophilic temperatures (20-45C). Using anaerobic thermophilic bacteria avoids both problems, however many genetic tools that have been originally developed for model organisms such as Saccharomyces cerevisiae and Escherichia coli do not work in these organisms. My group is working to develop several types of genetic tools necessary for domestication of thermophilic anaerobic bacteria, including:
- Ways to get foreign DNA into cells
- Tightly-controlled inducible promoters for reliable temporal control of gene expression
- Plasmid-based gene expression systems
- Chromosomal editing tools
- Ways to control the mutation rate
-
Metabolic pathway characterization and development
Metabolic pathway characterization and development
Metabolism can be understood at many levels of aggregation from individual enzymes to the whole organism. An important intermediate level of aggregation is the metabolic pathway. Developing pathways that enable rapid production of desired compounds at high yield and titer requires a detailed understanding of both the individual components and the systems-level behavior that results from the interaction of these components, including:
- Identification of constituent enzymes in a pathway and the stoichiometry of the reactions they mediate
- Characterization of enzyme inhibition and regulation
- Development of screens and selections to improve properties of key enzymes.
- Protein engineering to increase activity, decrease inhibition, or change substrate specificity
-
Physiology of native biomass-fermenting organisms
Physiology of native biomass-fermenting organisms
Currently, most ethanol is produced using various strains of yeast. These organisms are very good at producing ethanol but have no native ability to consume lignocellulose. This has made it difficult to develop cost-effective yeast-based processes for lignocellulosic ethanol production. My group takes an alternative approach of starting with organisms that natively ferment lignocellulosic biomass and engineering them for efficient biofuel formation. To do this, we need to understand key aspects of the physiology of these native biomass-fermenting organisms, including:
- Which substrates they can consume, and how these substrates are used for growth, energy production, and product formation.
- Factors that limit growth and fermentation.
- Genetic adaptation to stresses associated with industrial fermentation
Selected Publications
 Complete Works and Publications
Complete Works and PublicationsPatents
- Engineering an increase in ethanol production by altering co-factor specificity | 10767196
- Increased ethanol production by thermophilic microorganisms with deletion of individual HFS hydrogenase subunits | 10619172
- Engineering microorganisms to increase ethanol production by metabolic redirection | 9803221
Videos
Engineering Microbes for Cellulosic Biofuel Production (Seminar)
Suds & Science
News


